A few weeks ago a paper titled Life Before Earth was posted on the arXiv preprint repository. It came to my attention by way of this MIT Technology Review article and this blog post. The paper, using a rather simple extrapolation, argues that the apparent rate at which the complexity of terrestrial life increases suggests that its birth occurred approximately 9.7 billion years ago. Earth, in contrast, is around 4.5 billion years old. If their extrapolation is to be believed, then this discrepancy can only be resolved if terrestrial life is in fact of extraterrestrial origin. I will briefly summarize their argument, but I will not attempt to justify its validity. The original paper can be read here and is fairly accessible. The paper’s conclusions are consistent with a fact that has always puzzled me; the surprising complexity and maturity of what is known as the Last Universal Common Ancestor. It is this topic that I wish to focus on in this post.
First, let me recap their argument. They posit that a Moore’s law-like phenomenon governs biological complexity. In particular, by treating genome size as a proxy for complexity, and plotting the genome size of various organisms as a function of time, they obtain the following figure (from their paper):
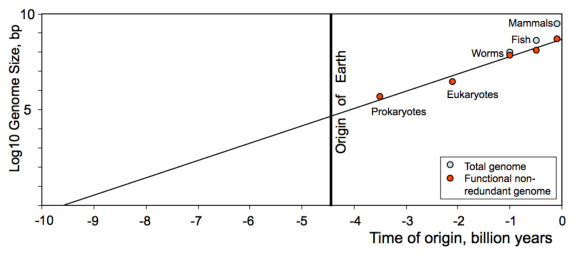
Empirically speaking genome size appears to be doubling every 376 million years. By extrapolating backwards in time to find the date when genomes were only a single basepair long, they conclude that the date is around 9.7 billion years ago, give or take 2.5 billion years. That is clearly outside the range of the Earth’s inception and is thus suggestive of panspermia, the theory that life arrived to Earth and was not conceived on it.
Although there are many caveats to the paper, the most glaring of which is the assumption of a constant rate of increase of genome size, I think it does possess a certain elegant simplicity that should not be dismissed out of hand. The paper is also consistent with a fact that I have always found puzzling and suggestive of panspermia.
For a moment, imagine that the “simplest” life forms on Earth were cats. No worms, no flies, no insects, no plants, and no bacteria. If, when working our way back through our evolutionary ancestry, we find that the tree of life suddenly stops. If the trunk, to stretch this analogy further, at which all life meets was thick and full of complexity, instead of being a thin hair-like stem. Well, while life does not stop at cats, the point at which all life meets is surprisingly complex, far more complex than what one could realistically expect to be the very first life forms. Before I go any further, let me make the discussion a little more precise since I have already stretched the analogies past their breaking point. To begin with, it is inaccurate to describe cats, or bacteria for that matter, as being earlier than we are on the evolutionary tree. All extant life is contemporaneous with humans. A more apt analogy is that of cousins. We are of the same evolutionary age as the rest of all terrestrial life. And it is quite clear, given the presence of life forms ranging in genome size from simple bacteria (~0.6Mbps) to lungfish (~133Gbps), that Earth is capable of simultaneously supporting a wide range of biological complexities, at least as measured by genome size. Thus the question is not so much about the distribution of extant genome sizes on Earth, but what this distribution, and specifically what the genomes themselves, tell us about the history of genomes on Earth. In particular, a process known as phylogenetic reconstruction allows us to take the genomes of two or more contemporary organisms, i.e. “cousins”, and ask what the genome of their common ancestor looked like, i.e. “grandparent”. It is this grandparent or ancestor that is of interest, because its hypothetical genome would have existed in the past, and thus can tell us something about the history and trajectory of genome evolution.
When attempting to phylogenetically reconstruct the ancestor of all life on Earth, what is known as the Last Universal Common Ancestor or LUCA, we discover several important things. For one, all life on Earth is related. This certainly did not have to be the case, but all three major domains of life (Archaea, Bacteria and Eukaryota) share a common ancestor. Furthermore, and this is the crux of my puzzlement, this ancestor is rather complex and full-featured. It is a lot like the “cat” in my initial example.
Briefly, here are some of the things we know about LUCA. First, it was basically a modern organism by most measures. It obeyed the basic dogma of DNA → RNA → proteins, it used the same set of amino acids and possibly genetic code as modern life, it had the same basic cellular structure and compartmentalization as modern bacteria, and it had a basically modern metabolic system. These facts in themselves are not necessarily unexpected. If there existed life forms that were fundamentally different, for example if they did not use nucleic acids, then it is likely that they would have been quickly marginalized by the “mainstream” strain of life and ultimately eliminated, which would explain why we do not see any of their descendants. One reason this would be so is because all life, and in particular bacterial life, benefit greatly from a constant exchange of genetic material. Any life form that was allowed to tap into this genetic pool would have an enormous evolutionary advantage over life forms that did not. Conversely, a life form that used genetic material that was incompatible with the “mainstream” strain of life would be excluded from the genetic pool, and thus be at a significant disadvantage.
The second thing we know about LUCA is that it had a surprisingly modern protein repertoire. Proteins are the machines of the cell, the molecules that actually do stuff like enable the cell to move. To give one example of the surprising modernity of LUCA, I will focus on helix-turn-helix (HTH) proteins, a class of proteins with which I am intimately familiar. These proteins carry out an important cellular function. They bind DNA and regulate which genes are synthesized at a given point in time and at a given cellular state. They, in essence, control the programming of the cell, its internal logical circuitry. All life on Earth has some set of proteins that carry out this function, although they do not all have to be of the HTH class. What is remarkable however is that LUCA contained not one or two HTH proteins, but at least 6 to 11 different types of HTHs. This represents a remarkable level of sophistication, and suggests that by the time LUCA emerged, life had a significant amount of time to evolve and settle on not just the basics of its biochemistry, but also the details of its more advanced functions.
Why is this so surprising? Let’s consider in a little more detail what it means for something to be in LUCA. The following schematic may give a useful mental image:
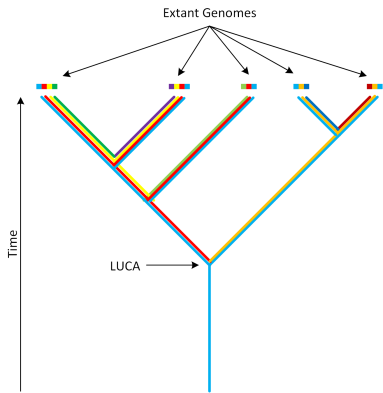
We start with extant organisms, the points at the top of the figure, and work our way back in time. Each organism is represented by colored blocks that signify its genetic makeup. For visual simplicity, we will assume that organisms only increase in complexity by adding genetic material (something that is untrue but is of no consequence to this discussion). The emergence of a new organism is visually represented by a split in the tree. Extant organisms have undergone many splits and so are comprised of many colored blocks. As we work our way back through time, we encounter the split points that denote two distinct organisms. The genetic material that is shared by these two organisms is represented by one color and that color persists further down the tree. By looking for the “colors” that are shared by all life on earth, we are able to genetically reconstruct what LUCA’s genome looked like. As a corollary of this, every branch of life that split off before LUCA must have hit a dead end. I.e. in reality we must have a situation that looks like this:
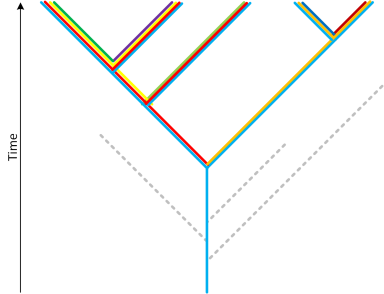
This is why LUCA’s genomic complexity is puzzling. It is reasonable to surmise that more fit organisms can outcompete and eliminate less fit ones, thereby destroying their evolutionary history. But what is surprising here is that every such branch, from before LUCA, must have been destroyed. This is by construction. If it were not so then we would have used it in the reconstruction of LUCA. And so what LUCA’s complexity is telling us is that any organism that was simpler than LUCA was sufficiently unfit that none of its descendants survived. This is a rather bold statement, and appears to me unsupported by the evidence. The argument I provided earlier, regarding access to the genetic pool, is inapplicable here. Losing a few HTHs would not have made the precursors of LUCA horribly unfit. And it is not just about HTHs. The overall protein repertoire of LUCA is rather impressive, and one that, as far as the known evidence suggests, is unlikely to have represented a qualitative leap in evolutionary fitness.
An extraterrestrial origin of terrestrial life, in which LUCA’s complexity came prepackaged, does offer a reasonable resolution to this puzzle.
As always, there are many caveats. First, as we discover more organisms, we will continue to refine our picture of LUCA, and it may ultimately end up being a lot simpler than we currently believe. Second, we may have simply missed a whole branch of life, or at least a set of organisms that fall within the current categorization but that are much harder to detect, and that happen to be much simpler. Basic and fundamental discoveries in biology continue to be made all the time; this is a young science. Finally, and perhaps most importantly, I am not an evolutionary biologist and so I only have a passing familiarity with the LUCA literature. I may very well have a missed an important and obvious point.
You use the term “modern” in a curious way. Wouldn’t it be more accurate to say that all current life forms have “ancient” proteins, biochemistry, etc. than that organisms long ago had a “modern” portfolio of these things? And, isn’t there evidence that complex organisms evolve quickly in the face of large changes in the environment (major warming or cooling, rise of the oxygenated atmosphere, etc). In fact, the rise of oxygen in the atmosphere itself might have led to a massive die-off of the preexisting organisms, hence erasing the prior record. In short, the assumption of a constant rate of evolution of complexity seems to me to be pretty dubious.
Fair enough point about the use of “modern”; I just meant to say that they appear largely unchanged, and to emphasize that it was their apparent modernity that is surprising. The assumption of a constant rate of evolution is irrelevant to my point. That was the assumption of the arXiv article.
A massive die-off type of scenario is possible, but something about LUCA must have made it uniquely able to adapt to those changes (this was way before oxygenation), such that anything simpler than LUCA could not have survived. It’s a possibility of course, but I don’t think anyone has found anything special about the properties of LUCA in that regard, at least not yet. In addition, this die-off must have hit everything other than LUCA and its descendants. I.e. all “cousins” of LUCA, anything that shared an ancestor with LUCA that is older than LUCA, must have been eliminated. This I find to be very unlikely, even in the case of a major die-off. Sure the historical record of many organisms could have been wiped out, but it must have happened in a way that is very selective, that eliminated anything that traces its ancestry further back than LUCA. One can argue that this is circular, that LUCA is simply what we get by phylogenetically reconstructing from everything that was not hit by the die-off. But then this brings us back to my original question. Why is it that LUCA ends up being so complex when we reconstruct it phylogenetically?
I study the evolution of lentiviruses. HIV-1 is just one of dozens of lentiviruses. The rate of evolution of lentiviruses has been accurately measured in thousands of isolates for the last 20+ years, and it is roughly 0.5% per year, such that viruses which shared a common ancestor one year ago (for example a blood donor whose blood was transfused into several recipients) are not 1% different from each other (each one changed 0.5%). So the naive conclusion is that after 100 years, the viruses will be 100% different. But any two random stings of DNA are 25% identical by chance because there are just 4 bases (A, C, G and T).
What we observe instead, is that the distance/time relationship is not linear. There are many sites in the genome that are mutating back and forth at a high rate A -> G -> A -> G … And other sites that are invariant over time.
HIV-1 is roughly 50% divergent from HIV-2, so early estimates were that HIV-1 and HIV-2 shared a common ancestor some 50 years ago. We now have solid evidence that their common ancestor was at least 500,000 years ago and much more likely to be closer to 5 million years ago.
The point is that inferring the ancient past from what we observe today is not always accurate. And genome size is likely not observed to be increasing in any linear manner even in recent times. The size of bacterial genomes is more likely to have decreased over time as they became more efficient, shedding “junk DNA” for example. And genome size among eukaryotes is not correlated with “complexity” of the organism. Mammals have smaller genomes than most plants, and genome size among single-celled eukayotes is highly variable, and often larger than the genomes of mammals.
These are all very reasonable comments although I do not think they impinge on the specific arguments in my post? With respect to the original arXiv article, they do not assume that genome size increases linearly. It most certainly does not. It is exponential and hence the appeal to Moore’s law. At least empirically speaking, given the organisms that they look at, exponential increase in genome size does appear to hold rather well (they’re looking at non-redundant genome content.)
I agree that there are many potential issues, and that phylogenetic reconstruction is dicey, particularly in bacteria/viruses which do not form a tree of life of any sort. It’s a matter of forming a hypothesis given the available evidence, with the caveat that said evidence is very shaky in both directions (terrestrial vs. extraterrestrial origins).
Hi, I took a biology class in high school and also watch Star Trek so I think I know what I’m talking about. If you watch http://en.wikipedia.org/wiki/The_Chase_(Star_Trek:_The_Next_Generation) you will know that indeed, our life had an extraterrestrial origin!
Haha yes I have seen that episode!
Entertaining read. As a side note, the implication of the paper would be exogenesis, rather than panspermia– a similar but technically different concept (as panspermia suggests that life is universal).
Howdy, I do think your site may be having browser compatibility problems.
Whenever I look at your web site in Safari, it looks fine
however when opening in Internet Explorer, it has some
overlapping issues. I merely wanted to give you a quick heads up!
Other than that, great website!
Thanks for the heads up! The layout is one of the standard WordPress themes, and so they must have not bothered to make it compatible with IE. I’ll see about contacting them about it.